Central Service Project: standardized biobanking, human probe and model system evaluation, data-base, bioinformatics
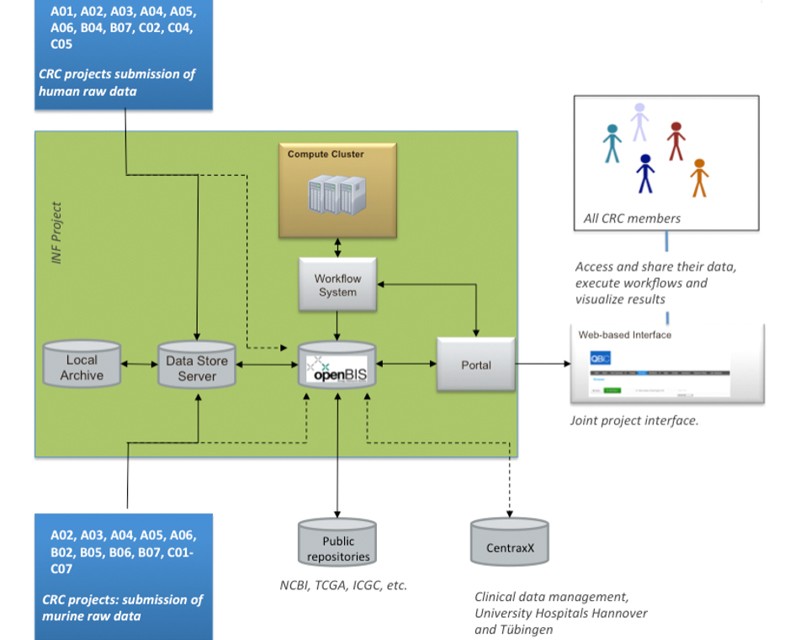
Central issues of the SFB/TR 209 are the quality of materials and models used for the research projects, availability of core data sets to all projects, optimisation of correlative data analyses, and sustainability of the generated results. The majority of projects within the collaborative research centre rely on human probes (70 % of the projects) or innovative mouse models of liver cancer (90 %) and intend to produce moderate to large amounts of hig-throughput data (75 %). The availability of appropriate samples and associated data, sound experimental design, and the consistent generation of high-quality data and metadata are keys for the project success. In addition, cross-project use of bioprobes and data analyses provides one of the main strengths and added value of the consortium. The Central Service Project INF provides three essential and expert-guided functions required for the success of the SFB/TR 209: a) Histotechnology and histopathological evaluation of human tissue probes and mouse model systems used in the different subprojects, biobanking, and harmonisation of cell culture activities (https://www.klinikum.uni-heidelberg.de/CMCP.142698.0.html), b) data management, database maintenance, and data sustainability, and c) experimental design, data integration/visualization, and bioinformatics support; Project INF will thus provide state-of-the-art infrastructure, work together with researchers from the various projects to plan and conduct the respective experiments, and support them with tailored probes and analyses for and across the projects.All three project components are based on long standing, successful, and sustainable infrastructures at each location and ask solely for add on funding required to cover the specific activities of the SFB/TR 209. Combination of all three project parts in one INF project provides synergies across the tasks and for complex data analyses.